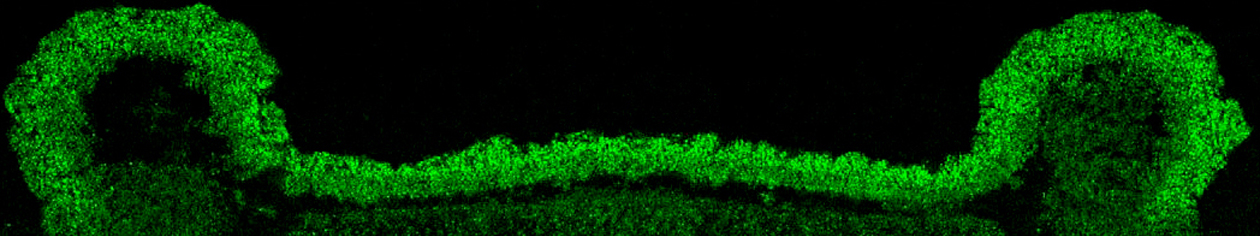
December 2014:
December 3-5, 2014, Gregor Gilfillan visited Prague for the first Steering Committee Meeting. The meeting was joined with scientific discussions and planning of the YEASTSEQ work flow.
March 2015:
Gregor Gilfillan visited Charles University in Prague for the two weeks (8th-21st March, 2015) to expand the knowledge and skills of the Czech team members, in accordance with the long-term cooperation aims of the YEASTSEQ project. Gregor Gilfillan taught PhD students and young researchers of CUNI and IMIC groups to set up the FAIRE and Chromatin Immunoprecipitation (ChIP) techniques, which will be used in the determination of epigenetic regulation involved in the reprogramming of structured biofilm colonies. Second Steering Committee Meeting was organized during the visit.


May 2015:
PhD student Jana Maršíková (from Faculty of Science, Charles University in Prague) attended training in Oslo University Hospital sequencing facilities from 11th to 15th May 2015 to learn sample preparation and library constructions for next-generation RNA sequencing. RNA samples from six different types of differentiated cells of yeast colonies were taken by Jana from Prague to Oslo and libraries were prepared during her training. The training was provided by Gregor Gilfillan and the knowledge gained will be cascaded to other CUNI and IMIC group members in Prague.

Click here to see more pictures from Oslo.
June 2015:
Bioinformatician Alexandru Mizeranschi visited Oslo from 9-26 June, 2015 to receive training by Timothy Hughes in bioinformatics tools needed in YEASTSEQ project. The training focused on the use of bioinformatics software for the analysis of next-generation sequencing data, produced from RNA-Seq, ChIP-Seq and DNA sequencing experiments. During the visit, Alexandru also set up an Amazon EC2 cloud server that is used for storing and analyzing the sequencing data of YEASTSEQ.
June 2015:
June 17-21, 2015, Zdena Palkova and Libuse Vachova visited Oslo for the third Steering Committee Meeting. The meeting was joined with scientific discussions on the experimental progress and planning of the future YEASTSEQ work flow.

Click here to see more pictures.
November 2015
Otakar Hlaváček (junior scientist, Institute of Microbiology) visited Oslo University Hospital sequencing facilities from 24th to 28th November 2015 to learn DNA sample preparation and library construction using PCR‑free protocol for TrueSeq sequencing technology. DNA samples from 14 different sectors of yeast colonies and 5 control samples were taken from Prague to Oslo and libraries were prepared during his training. The training was provided by Gregor Gilfillan and the knowledge gained will be provided to other CUNI and IMIC group members in Prague.

March 2016
Gregor Gilfillan visited Prague (29th Feb – March 2nd) for the 4th steering committee meeting. Additional scientific discussions covered RNA-seq data analysis and the FAIRE method (see pictures). Lunch at Pivovarsky dum, a favourite nearby restaurant – in the expectation that this was probably the last meeting before the Prague labs relocate to their new institute in the South of the city.


June 2016:
June 18-21, 2016, Zdena Palková and Libuše Váchová visited Oslo for the fifth Steering Committee Meeting. The meeting was joined with scientific discussions on the experimental progress and planning of the future FAIRE and ChIP experiments.
September 2016:
September 25-29, 2016, David Segorbe, postdoctoral fellow from the Charles University, traveled to Oslo in order to receive training into create DNA libraries using Faire samples that he previously produced in Prague. He was trained and supervised by Dr, Gregor Gilfillan.
November 2016:
November 8th, 2016, Gregor Gilfillan visited Prague and attended with the group leaders Z. Palková and L. Váchová The 2nd Czech-Norwegian Research Conference – Prague 2016.
On the next day, November 9th , 2016, Gregor Gilfillan attended the 6th steering committee meeting. A possible future collaboration was discussed based on the use of phenotypic switching as an epigenetic toxicology test.
December 2016
Derek Wilkinson, post-doc from the Faculty of Science, Charles University, attended the course in proteomics bioinformatics organized by the European Bioinformatics Institute (EMBL/EBI) at the Wellcome Genome Campus at Hinxton, near Cambridge in the UK. The course provided hands-on training in the basics of mass spectrometry and proteomics bioinformatics, search engines and post-processing software, quantitative approaches, MS data repositories, the use of public databases for protein analysis, annotation of subsequent protein lists and incorporation of information from molecular interaction and pathway databases.